This is the final post in a three-part series (I strongly suggest reading part I and part II before continuing) describing my attempt to identify “safe” and “unsafe” foods for Inflammatory Bowel Disease (IBD; including Ulcerative Colitis and Crohn’s Disease) using various machine learning techniques and Python. Part III below walks through the results of this stage of the study, conclusions, and future work. The working project is available at https://github.com/ZaxR/ibd_diet_analysis and a [reliable] working view of the jupyter notebook is available via nbviewer.
Results
Using association rule learning (fp-growth) and a custom cross-validation technique (not typically used since inherently association rule learning is self-validating) an average of 18-19 new food recommendations were able to be made to survey participants, with 80%+ accuracy (usually 90%+). Importantly, regardless of reasonable tweaking of support and confidence thresholds, results were accurate (in line with or higher than the chosen confidence levels) and valid (no commonly occurring incorrect answers) across multiple tests.
However, there was a significant percentage of missed recommendations (65% at peak in the test range) and very few new recommendations (<2 at trough in the test range) being made at higher support (15%+) and confidence (90%+) levels. If the model can’t make many new recommendations to users, the model will be minimally useful at best (at least until more data is collected). As a result, the fp-growth parameters were tweaked to a support of 10% and a confidence of 80%, which results in only 35-40% of recommendations being missed and 18-19 recommendations being made on average, while accuracy and validity are maintained (across 10 sample runs). While a drastic improvement, those stats still reflect only 20-25% of possible recommendations (improved with the final model) – a solid, but not as-good-as-hoped result (and a result that will influence future work; more below).
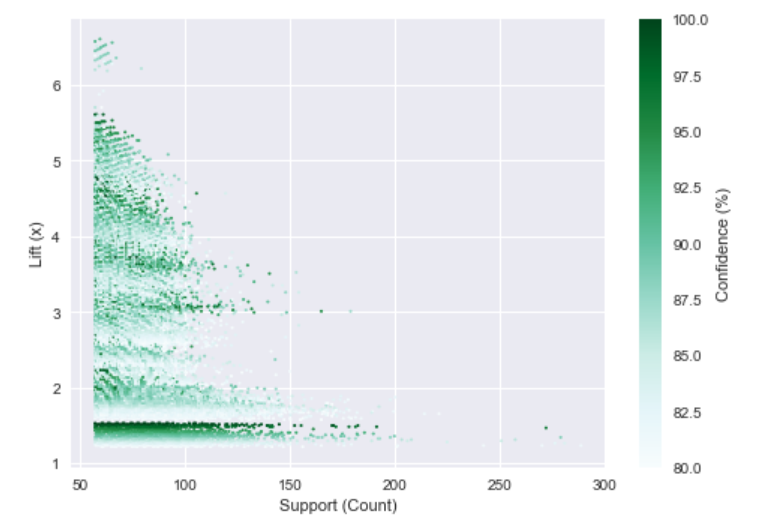
Using all of the data to build a “final” model, 888,926 rules were generated (using thresholds of 10%, 80%, and 1x for support, confidence, and lift, respectively) with rules being generated for 74% of possible recommendations. The two figures below reflect a summary of the generated rules.
To summarize more loosely, here are a sampling of some rules:
- Can eat rules: animals (chicken, beef, various fish, pork), white rice, potatoes, and ‘staple’ vegetables (carrots, cucumber, lettuce, tomato).
- Can’t eat rules: apple juice, coffee, cola, raisins
- Rules common in training sets, but didn’t quite make the final parameter cut: not alcohol of various types
- Commonly conflicting rules: fruit, dairy, cruciferous vegetables
Among the most common food recommendations (based on multiple train/test groupings are leeks (~36% of the time), lettuce (~34%), garlic (~33%), honeydew melon (~32%), cod (~32%), cantaloupe (~31%), chicken eggs (~31%), basil (~30%), cucumber (~30%), white potatoes (~30%).
Conclusions
Based on the results, clearly there are underlying patterns in foods IBD sufferers believe they can/can’t eat. While survey data alone can’t determine if there’s truly a medical impact or what such mechanisms would cause those reactions, the results suggest that there are some underlying relationships worth investigating.
However, it’s possible that survey answers are biased beyond reasonable use, given the complexity of assessing reactions to individual foods, various starting states of the IDB sufferers (in flare or not in flare), and influence from the various “fad” or “suggested” diets survey participants may be influenced by. That said, given the initial success with the developed model, at least as a proof of concept, and discoveries about the usefulness of the data set during data exploration, further analysis appears to be warranted after some adjustments to the survey.
Recommendations for Survey:
- Track continuous (floating) data as opposed to the current integer data, which would be relatively trivial given that the survey questions are already presented as a label-less sliding scale.
- Distinguish between whether a food can be eaten in some amount or sometimes (two distinct possibilities).
- More clearly defined food categories by eliminating the few overlapping food groups that exist (e.g. peppers (capsicum) and peppers, chili) and adding pictures of each food for users’ sake.
- Track changes in user answers over time if the ability to change answer is allowed.
Future work
While this marks the final post in this three part series for the ChiPy mentorship program, I plan to continue work identifying safe/unsafe foods for IBD patients, and patterns that may exist within those foods. I plan to improve and expand the work done using our initial model, both by hopefully having the survey improvement recommendations implemented and also by gaining access to the full survey participant information (including such additional information as type of IBD, medications, demographics, etc.). Additionally, it is planned to spend more time with the USDA nutrient database tool that was created during the project, looking for common vitamin/mineral/macro nutrient elements among foods recommended to users. Should more data become available (or when enough data using the new float data is collected), investigating more advanced and scalable models (such as neural networks) could help improve the recommendations/other learnings from the data.
Separately, given the success of the model to-date, working with ibdrelief.com to create an API and build a front-end recommendation tool for survey participants, with a live “learning” model with feedback is also on the roadmap.
Ultimately the hope remains that learnings from the analysis of available survey data will ultimately help users eat confidently and help direct more targeted medical research in an effort to cure IBD.
Special thanks
Special thanks to my mentor, Ed Gross, for his continual project guidance (despite it being the final weeks before his wedding!) and brilliant idea to use a logical ternary operator for the association model; and to my unofficial data science mentors, Chris Gruber (who helped me on top of having his own ChiPy mentee), Andra Stanciu, and Laurel Ruhlen. Without the guidance of each of them, I wouldn’t have learned nearly as much along the way, and certainly would have spent more time [idiomatically] banging my head against the wall.
And of course, thank you to ChiPy for making this entire program happen. If you live in Chicago, I strongly recommend getting involved with this program either by applying for a mentee slot or a mentor (we always need more) and/or check out the monthly meetup/speakers.